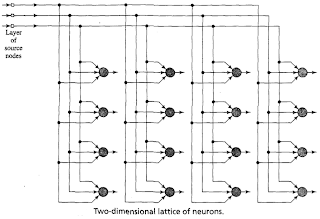
Notice: For an update tutorial on how to use minisom refere to the examples in the official documentation.The Self Organizing Maps (SOM), also known as Kohonen maps, are a type of Artificial Neural Networks able to convert complex, nonlinear statistical relationships between high-dimensional data items into simple geometric relationships on a low-dimensional display. In a SOM the neurons are organized in a bidimensional lattice and each neuron is fully connected to all the source nodes in the input layer. An illustration of the SOM by Haykin (1999) is the following
Each neuron n has a vector wn of weights associated. The process for training a SOM involves stepping through several training iteration until the item in your dataset are learnt by the SOM. For each pattern x one neuron n will "win" (which means that wn is the weights vector more similar to x) and this winning neuron will have its weights adjusted so that it will have a stronger response to the input the next time it sees it (which means that the distance between x and wn will be smaller). As different neurons win for different patterns, their ability to recognize that particular pattern will increase. The training algorithm can be summarized as follows:
- Initialize the weights of each neuron.
- Initialize t = 0
- Randomly pick an input x from the dataset
- Determine the winning neuron i as the neuron such that
- Adapt the weights of each neuron n according to the following rule
- Increment t by 1
- if t < tmax go to step 3
MiniSom is a minimalistic and Numpy based implementation of the SOM. I made it during the experiments for my thesis in order to have fully hackable SOM algorithm and lately I decided to release it on GitHub. The next part of this post will show how to train MiniSom on the Iris Dataset and how to visualize the result. The first step is to import and normalize the data:
from numpy import genfromtxt,array,linalg,zeros,apply_along_axis
# reading the iris dataset in the csv format
# (downloaded from http://aima.cs.berkeley.edu/data/iris.csv)
data = genfromtxt('iris.csv', delimiter=',',usecols=(0,1,2,3))
# normalization to unity of each pattern in the data
data = apply_along_axis(lambda x: x/linalg.norm(x),1,data)
The snippet above reads the dataset from a CSV and creates a matrix where each row corresponds to a pattern. In this case, we have that each pattern has 4 dimensions. (Note that only the first 4 columns of the file are used because the fifth column contains the labels). The training process can be started as follows:
from minisom import MiniSom
### Initialization and training ###
som = MiniSom(7,7,4,sigma=1.0,learning_rate=0.5)
som.random_weights_init(data)
print("Training...")
som.train_random(data,100) # training with 100 iterations
print("\n...ready!")
Now we have a 7-by-7 SOM trained on our dataset. MiniSom uses a Gaussian as neighborhood function and its initial spread is specified with the parameter sigma. While with the parameter learning_rate we can specify the initial learning rate. The training algorithm implemented decreases both parameters as training progresses. This allows rapid initial training of the neural network that is then "fine tuned" as training progresses.
To visualize the result of the training we can plot the average distance map of the weights on the map and the coordinates of the associated winning neuron for each patter:
from pylab import plot,axis,show,pcolor,colorbar,bone
bone()
pcolor(som.distance_map().T) # distance map as background
colorbar()
# loading the labels
target = genfromtxt('iris.csv',
delimiter=',',usecols=(4),dtype=str)
t = zeros(len(target),dtype=int)
t[target == 'setosa'] = 0
t[target == 'versicolor'] = 1
t[target == 'virginica'] = 2
# use different colors and markers for each label
markers = ['o','s','D']
colors = ['r','g','b']
for cnt,xx in enumerate(data):
w = som.winner(xx) # getting the winner
# palce a marker on the winning position for the sample xx
plot(w[0]+.5,w[1]+.5,markers[t[cnt]],markerfacecolor='None',
markeredgecolor=colors[t[cnt]],markersize=12,markeredgewidth=2)
axis([0,som.weights.shape[0],0,som.weights.shape[1]])
show() # show the figure
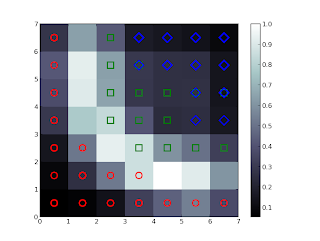
The result should be like the following:
For each pattern in the dataset the corresponding winning neuron have been marked. Each type of marker represents a class of the iris data ( the classes are setosa, versicolor and virginica and they are respectively represented with red, green and blue colors). The average distance map of the weights is used as background (the values are showed in the colorbar on the right). As expected from previous studies on this dataset, the patterns are grouped according to the class they belong and a small fraction of Iris virginica is mixed with Iris versicolor.
For a more detailed explanation of the SOM algorithm you can look at its inventor's paper.



Hey, i've been using your MiniSOM, it works great..
ReplyDeletei want to ask, how can we save the trained model for future use? so we dont have do the train again. Thanks in advance :)
Hi Guntur, MiniSom has the attribute weights, it's a numpy matrix that can be save with the function numpy.save. If you want to reuse a MiniSom, you can crate an instance, train and save the weights. When you want to reuse that network, you can create a new instance, load the weights from a file and overwrite the attribute weights.
Deletethanks for the quick reply.. i will try that :)
Deletecan we plot confusion matrix using the trained model.
Deletehi rebelgirl, you can look at this example https://github.com/JustGlowing/minisom/blob/master/examples/Classification.ipynb
DeleteYou have to use the appropriate sklearn function to compute the confusion matrix.
This comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteHow does the SOM react to categorical inputs, e.g. [1,2,3,..] are categorized peoples favorite foods? Is there a way to work with this type of data? Does the training assume an ordinal number set or does it learn this on its own?
ReplyDeleteThe algorithm assumes that the order of the values has a meaning. I suggest you to try a binary encoder like this one: http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.LabelBinarizer.html#sklearn.preprocessing.LabelBinarizer
DeleteHello.
ReplyDeleteThis looks like a promising library.
I noted you used a 2D map, 7 by 7, for output neurons. Is 3D map also possible? Could you please illustrate this if possible.
Thanks
Hi John, I'm sorry but this library supports only 2D maps at the moment and I don't plan to extend it at the moment.
DeleteHello
DeleteThanks for the information
This comment has been removed by the author.
ReplyDeleteHi Walter,
DeleteI can't read your data type but I'm pretty sure that the problem is in the shape of matrix that you are using. Please check that it has no more than 2 dimensions.
Hi Walter,
ReplyDeleteHi can I install the package into my ubuntu via terminal.
yes, you can. Check the documentation: https://github.com/JustGlowing/minisom
DeleteThanks Just Glowing,
DeleteI have gone through your github account before.
I am unable to identify despite my effort.
Please guide me with the 'pip install' command for the installation in ubuntu.
Please share the link to down load MiniSom for ubuntu.
DeleteIf you want to use pip, you just need to run this command in your terminal:
Deletepip install git+https://github.com/JustGlowing/minisom
Hello
ReplyDeleteFrom the above illustration on iris, suppose we have a flower with the following measurements: 20,8,16 and 7. How can one use the trained SOM to determine the class of this new flower/data?
Thanks
You can get the position on the map using the method winner and assign the class according to the class of the samples mapped in the same (or close) position.
DeleteThanks
DeleteI will try it out
hello
ReplyDeleteI am using your MiniSom for anomaly detection in high dimentional data but I have a problem with the choice of the input parameters
So how can we chose these parameters using the dimension of the input data?
Hi, I covered parameters selection in a different post:
Deletehttp://glowingpython.blogspot.co.uk/2014/04/parameters-selection-with-cross.html
Hi, I am using your MiniSom for abnormaly detection in a data stocked in Elasticsearch, so how to recover my data in your code ? and how to replace your iris.csv with my data? should i convert my data to numpy matrix? if yes how?
ReplyDeleteHi pikita,if you want to parse a csv I recommend you to use pandas. You'll find the function read_csv pretty useful.
Deletehi, thank u for your answer but i didn't understand how to use pandas have u a link ?
DeleteCheck this out: http://chrisalbon.com/python/pandas_dataframe_importing_csv.html
Deletei have all my data in elasticsearch i don t know how to recover specially the code I want in your code, and then convert it to a numpy matrix ??
Delete?
DeleteHi, How to identify the accuracy in this code i.e. how many sample classified as correctly in each class
ReplyDeleteHi Naveen, no classification is done here.
DeleteFantastic piece of code, thanks for sharing it. I've been playing with it to classify oceanography data (pacific ocean temperatures) and am wondering about reproducibility. While it picks up the obvious patterns (i.e. El Nino) it seems that each time I run the algorithm I get different maps, sometimes patterns a show up as maps and other times are not present, and increasing the number of iterations doesn't seem to cause a convergence to consistent results. This is also true for your supplied iris and colors examples. Am I missing something, or do SOMs truly provide different results each time the algorithm is run?
ReplyDeleteHi Eric, the result of the algorithm strongly depends on the initialization of the weights and they are randomly initialized. To have always the same results you can pass the argument random_seed to the constructor of MiniSom. If the seed is always the same, the weights will be initialized always the same way., hence you'll have always the same results.
DeleteNice work.I have gone through your code,and I just wonder that why 'som.random_weights_init(data)' is called,for the weights already have been initialized in the MiniSom __init__ function。And 'som.random_weights_init(data)' regards normaliztion of the data as weights,is it a correct way?Also 'som.random_weights_init(data)' replaced the initial weights.
ReplyDeleteHi, som.random_weights initializes the weights picking samples from data. This is supposed to speed up the convergence.
DeleteThanks for your reply.
DeleteI excerpted your code below:
self.weights[it.multi_index] = data[self.random_generator.randint(len(data))]
self.weights[it.multi_index] = self.weights[it.multi_index]/fast_norm(self.weights[it.multi_index])
In the function,we iterate the activation_map,the first line means picking samples from data,and assigned to weights.That's to say,we swept away the initialization in Minisom's __init__ founction.Why so?
Because that initialization seemed more suitable in that case. The constructor initializes the weights using np.random.rand by default. It's totally fine to choose a different initialization if the dafault one doesn't suits you.
DeleteThank you.That means both __init__ and random_weights_init are fine? And we can choose one of them or define a newer one.
Deleteyup
DeleteHi, How can I plot the contour of neighborhood for each cluster. The weights matrix correspond to the U-matrix?
ReplyDeleteHi Mayra, you can compute the U-matrix with the method distance_map and you can plot it using pcolor.
DeleteHi, thank for answer my question. I have a doubt. I was testing your code with the mnist data set, is similar to the digits dataset from python, but the difference is the size of the images. I trained the network SOM with a sample of 225 random digits and the dimension of my grid is 15*15. When I plot the U-matrix with the method distance_map, each coordinate of my plot should have a digit rigth? Why the position of my winning neuron is the same for different samples? There are empty positions in muy plot wuth any digits. When training the SOM with a larger sample there are empty positions too in muy plot. Should that happen? I use other implementation for example kohonen library from R package does not happen. Could you help me understand why this happens?. Thanks
ReplyDeleteHi again,
DeleteI'll try to answer our questions one by one.
- If your samples represent digits, you can associate a cell in the map with a digit. See this example https://github.com/JustGlowing/minisom/blob/master/examples/example_digits.py
- If two samples have the same winning neuron, it means that the two samples are similar.
- With the training and initialization methods implemented, It's normal that some areas don't have winning neurons for the samples used for training, especially between regions that activate for samples that have different.
Thank you so much for providing the code. I hope you can help me with my question. I thought that it is possible to apply SOM for outlier detection in an unsupervised manner (without labeled data). In the iris dataset the data has labels right:
ReplyDeletet = zeros(len(target),dtype=int)
t[target == 'setosa'] = 0
t[target == 'versicolor'] = 1
t[target == 'virginica'] = 2
If I just have numerical data without any labels, how can I use your SOM approach?
Thank you very, very much for your help! I really appreciate it! :-)
Nevermind! Got it! Thanks a lot! :-)
DeleteBut how can I count the amount of samples in one position?
DeleteHi, look at the documentation of the method win_map, I'm sure that will answer your question.
DeleteHi, that's a great library you have implemented. I would like to try and combine a self organising map and a multilayer perceptron. I have used your network to cluster character images. Is there any way to save the clustering so I can feed it as input to a multilayer perceptron?
ReplyDeleteHi, you need to use the method winner on each sample in your dataset and save the result in a format accepted by your MLP implementation.
DeleteHi, thankyou fot the fast response. I shall try this, thank you.
DeleteI have labels associated with each image in the dataset very similar to t in your example. I want to associate the winner with a label if possible. When you use a for loop and use the enumerate function on the data, is the code t[cnt] associating your t label with the winner?
Yes, with t[cnt] the label of the sample is considered to assign a color to the corresponding marker.
DeleteIt will be very helpful if u provide the source code for classification and dimensionality reduction of images(not numericals) using SOM and store that result in a format accepted by general MLP.
DeleteHi,
ReplyDeletethank you for this great library.
When we find the outliers on the map by looking at the highest distance (close to 1), how can we know to which observation in the original data it corresponds ? In other words, is there an inverse function of the winner() function to reverse the mapping from the input space to the output map ?
Thanks for your help.
hi, winner() can't be inverted, but win_map() will solve your problem.
DeleteThank you very much for your reply.
DeleteHi, do you have any function to calculate the SOM-MQE? Thanks!
ReplyDeleteHi, I'm not sure this is exactly what you need but can help you: https://github.com/JustGlowing/minisom/blob/master/minisom.py#L167
DeleteHi, is there a way to change the "lattice", "shape" and "neigh" of the map? Thanks!
ReplyDeleteHi Jianshe, there's no way to do that without changing the code currently. The philosophy of the project is to be minimal.
DeleteHi JustGlowing,
DeleteThanks. The reason why I am asking this is that the clustering results and quantization errors are different from the results I got from MATLAB somtoolbox. After reviewing all the codes inside, I think the possible reason maybe the parameter settings on the training stage. After all, I think you did a brilliant job. Thanks!
Hi,
ReplyDeleteI am having a trouble with your pylab code from your example digits.py which I found here: https://github.com/JustGlowing/minisom/blob/master/examples/example_digits.py
I have digits images I want to plot but the problem is a window titled figure 1 comes up with an axis all by itself and a separate window titled figure 2 pops up with the clustered images but without an axis. I tried to combine the 2 figures by removing figure(2) from the code and even though only one window titled figure 1 pops up there is no axis, just the clusteted images alone. On top of that not all of the clustered images are shown up. Do you know of anyone else having this problem who fixed it or if there is a way to combine the 2 figures or a way for it the axis and clustered images to show up using 1 figure?
Thanks
Hi, first congratulations for your work and thanks for share it!
ReplyDeleteI am trying to implement MiniSom for a 3D array. The array´s shape is [time(days),latitude,longitude] and the intention is to catalogue each day in one of four nodes. For the moment I am reshaping it to an array with the shape [time(days),len(latitude)*len(longitude)]. This way:
som = MiniSom(2,2,len(latitude)*len(longitude),sigma=1.0,learning_rate=0.5)
som.random_weights_init([time(days),len(latitude)*len(longitude)])
However, I believe it is not correct, as with the reshaping the relation between coordinates is lost. Am I correct? Can I implement in some way?
Thanks
Hi, this way the relation between coordinates is indeed lost. It's hard to give you a suggestion without knowing why you can't keep the original shape [time(days),latitude,longitude].
DeleteThanks for the quick reply!
ReplyDeleteI would like to keep the original shape, but I do not know how to implement with miniSom. In the documentation (https://github.com/JustGlowing/minisom) is indicated that "you need your data organized as a Numpy matrix where each row corresponds to an observation or an as list of lists like the following", but my problem is that I have a 2-D array instead of a list/row. Therefore I do not know what to introduce as "input_len" in the class MiniSom or if train_random assimilates 3-D arrays...
Thank you again!
Hi, if you have a 2D array you have to consider each row as a sample and each column as a variable. The parameter input_len is the number of columns in your array.
DeleteI am sorry for having explained it badly and for the insistence, but my matrix is 3D. Would it be possible in this case? How would be the input_len parameter?
DeleteThis comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteHi,
ReplyDeleteIs there a way to get the coordinate of the grid by seting a value range of the colorbar without looking at the map? For example, if I want to get the coordinate of the grid with the value between 0.9 and 1.0 on the colorbar.
Hi, the color of the grid is given by the matrix returned by distance_map(). You just have to check the elements in this matrix that are in the range you want to check.
DeleteHi, JustGlowing
DeleteThank you very much for your quick response.
This comment has been removed by the author.
ReplyDelete
ReplyDeleteHi
Thank you very much for this great library.
How can I cluster and assign the cluster number to samples?
How can I identify to which cluster new sample belongs?
Thank you.
Best regards
Hi © Илья Расин!
ReplyDeleteEach neuron represents a cluster, hence the position of the neuron that map you sample represents the cluster it belongs to.
Hi
DeleteThank you!
Regards
Hi,
ReplyDeleteThank you very much for your code. Would you please tell me how to overwrite the attribute weights with the saved weights?
Hi, the weights are stored in the attribute _weights as numpy matrix. You just have to overwrite it with your own matrix.
DeleteHi, JustGlowing
DeleteThank you so much for your quick reply.
Hi, JustGlowing
ReplyDeleteThank you for the code. Would you please explain to me what sigma does? You did mention that sigma is the initial spread, but I don't understand what that mean.
Hi, sigma is the initial spread of the neighborhood function. In case sigma is very small, only a neuron (the winning one) will be trained at each iteration. In case it's very big all the neurons will be trained.
DeleteHi, JustGlowing
DeleteThank you very much for your reply. That is very helpful.
Hi,
ReplyDeleteDoes the train_random algorithm gets finished once the SOM converges or does it keep going until it reaches the max iteration?
Hi, sorry for the late reply. The algorithm stops when the total number of iterations is reached.
DeleteHi,
ReplyDeleteThank you very much for your code.Could you please tell me if is there any way to identify the occurrence of topological distortions (when the network folds over itself)?
Like in this link: https://www.researchgate.net/figure/SOM-output-lattice-topology-distortion-examples-related-to-edge-folding-top-left_fig15_304024726
Hi. Is there a way to assign labels to markers? For example, get the Iris species categories to be visible on the SOM?
ReplyDeleteHi, have a look at this notebook: https://github.com/JustGlowing/minisom/blob/master/examples/examples.ipynb
DeleteYou need the latest version of minisom to run it.
Hi,
ReplyDeleteGreat work.
Is there a way to implement SOM on raw text corpus without any labels, to cluster on its own?
Of course, you could use one of vectorizers in sklearn to preprocess your documents and then run minisom on the data. This is a good vectorizer: sklearn.feature_extraction.text.TfidfVectorizer
DeleteThanks for the quick reply. I used the same, but it is only creating the single cluster (including all tokens in single cluster). Am I making mistake anywhere? Can you please share your thoughts on the same?
DeleteHard to say what the problem is. Make sure your data is normalized.
DeleteYes, the data is normalized. Steps I followed is that I vectorized the tokens, used train_batch to train SOM and trying to visualize. Am I making a mistake in visualization? Or can you help me on how can I get the number of clusters created by SOM or visualize the same. It will be really helpful. Thanks.
DeleteIf you are using SOM for clustering purposes, recall that each cell of the map can be considered a cluster itself. For example a 2-by-2 map, can reflect 4 clusters.
Delete(downloaded from http://aima.cs.berkeley.edu/data/iris.csv)
ReplyDeletethis link is not working so ,is there any another alternate to download that....
hi, you can find the dataset here: https://github.com/JustGlowing/minisom/blob/master/examples/iris.csv
Deletehi, think you for this great job
ReplyDeleteI want to get for every object the cluster or clusters relate to
think ones again
Hi, firstly thanks for this great work.
ReplyDeleteI have plotted the results of the SOM using the distance_map function. However, I now want to cluster similar neurons together. Am I correct in thinking that the distance_map represents the average distance of each neuron to its neighbourhood? Is there a way to get actual distances of each neuron so I could cluster neurons close to each other? Thanks
Hi, JustGlowing
DeleteI figured it out. Sorry for the trouble. Thank you.
Hi, Thank you for your wonderful work
ReplyDeleteI was looking at the code trying to figure out what the distance_map() was doing, but I couldn't understand it completely. Is it possible that you can give me some examples to help me understand it so I will be able to code it without copying your code. Thank you.
Hi, the distance map is nothing else than a U-Matrix: https://en.wikipedia.org/wiki/U-matrix
DeleteHi there.
ReplyDeleteThanks for sharing MiniSom.
I was wondering how could I get the component plane (plots) from the training?
(thought about it as the winner f(), but need to check.)
cheers
hi, check out this example: https://github.com/JustGlowing/minisom/blob/master/examples/DemocracyIndex.ipynb
DeleteThanks mate. cheers
DeleteI'm wondering if only having 100 iterations means that not all samples get used as inputs in this example? If I understand your code correctly, each time an input (or sample/row) is passed to the network/map, it counts as 1 iteration. Is understanding correct?
ReplyDeleteThis has been a great implementation btw. Thank you.
Hi there! Yes, in each iteration a single sample is processed.
DeleteGreat. Thank you! So in your tutorial, the last 50 samples are not used in training?
DeleteAll samples are used for training.
DeleteOkay, I am definitely confused now. In your tutorial you do only to 100 iterations in the training stage. There are 150 input patterns. If each iteration involves just one input pattern, the shouldn't there be 50 left over that aren't used for training?
DeleteYou are right, 100 iterations are used for training which means that the some only sees 100 samples.The entire dataset is used for the visualization.
DeleteCool. The result in the tutorial looks quite a bit better than the one I get. I know the result is somewhat random each time you train the SOM. However, this does not seem to be the cause of the difference.
DeleteI can get my result to look similar to yours if I increase the number of iterations to one or ten thousand (rather than just 100). Does the result in your tutorial come from only 100 iterations, or did you actually train on many more examples for the image you show in the tutorial?
You get different results because minisom has changed a lot during the years. You can find updated examples here: https://github.com/JustGlowing/minisom/
DeleteYou can get this results with 100, but only if the parameters are properly optimized. The neighborhood functions, sigma and learning rate changed massively since I did this example.
Oh really? That's really handy to know. The map I get with the parameters in the tutorial is not beautiful, haha. I suppose I will find the best parameters in your examples on Github. Thank you.
DeleteIs finding the optimal parameters just a matter of trial and error?
Parameters optimization is a broad topic.
DeleteAs I am discovering :)
DeleteThanks again, and happy new year
hi. im very interested with the application of ksom in solving clustering problems. i have a dataset of 2634 observations and 46 variables for my project. im using clustering.ipynb as my reference. i got error "scatter() got multiple values for argument 'alpha'" when trying to visualize it. how do i solve the error? thank you
ReplyDeleteHi there, just use the parameter alpha only once when you call scattered.
Deletethank you for the solution! it would be very helpful if you can visualize clustering using MNIST dataset?
DeleteHi, check this out: https://github.com/JustGlowing/minisom/blob/master/examples/HandwrittenDigits.ipynb
DeleteHello, I wanted to use this lib for unsupervised anomaly detection, can you please guide me how to understand the code ? how to detect anomaly or outlier from the given data set ? how to get the index of the anomaly point or row (which function is using for that)? how to get the final assigned labels after training ? Please guide me
ReplyDeleteHi, have you had a look at the examples directory?
Deletehttps://github.com/JustGlowing/minisom/tree/master/examples
There's one example specific for outliers detection.
yes, but not getting how to get the row or index of the anomaly
DeleteThe example shows you how to know if a sample is an outlier. Once you know that you can use the method som.winner to know its position on the map.
Deletethanks a lot
Deletehello. Thanks for this great library. I was wondering if there is a way to retrieve for each node the distance to all neighbor nodes. If I understand correctly, the distance_map() returns the average distance to all neighbors.
ReplyDeleteHi,
ReplyDeletehow to plot hexagonal Heatmap with this.
Please guide
Hi,
ReplyDeleteI saw a minor mistake in your code. Maybe you want to filter it out.
In the last code snippet you mention:
axis([0,som.weights.shape[0],0,som.weights.shape[1]])
However, I think it needs to be:
axis([0,som._weights.shape[0],0,som._weights.shape[1]])
I love your work! Really useful for me. I am also doing my thesis about this subject :) Trying to programm a Dynamically Controlled Network. So instead of decreasing your neigborhood function in time, we create a separate algorithm based on process analysis of the network itself to decide if the neigborhood decreases or not. Same holds for the learning rate. It is called DyCoN. Very interesting stuff.
Thanks again for your work!
Hi! Thanks for reporting that.
DeleteYou can find updated examples about how to use minisom here: https://github.com/JustGlowing/minisom/tree/master/examples
DyCoN seems very. I'll read something about it as soon as I have some time.
Good luck for your studies :-)
Hi, thank you so much for your great work.
ReplyDeleteSOM can be used as a dimensionality reduction method and a feature extraction method.
For the dimensionality reduction, input data dimension is N, number of neuron at the output layer is M. If M < N, then does it mean the dimensionality reduction?
For the feature extraction, are the weight matrix new features? But the size of this weight matrix is N*M, which is much larger than input data dimension. If I want to use SOM as a feature extraction method, what are the new features? Many thanks
Hi there, the answer to your first question si yes. To the second, you can use the position of the winning neurons on the map as features.
DeleteThank you for your reply.
DeleteFor my first question, the original iris dataset has 4 features, but SOM model's output size is 7*7=49 as your example above. So, for this particular problem, SOM is not for dimensionality reduction,right? But I can visualize 4-dimensional features(even higher dimensions for other datasets) into 2D map using SOM (very similar to PCA). I am very confused.
For my second question, if I use the position of the winning neurons as my extracted new features, the size of the new features is 2. Because they are location information, I don't know if these 2 new features have information loss. Do you have any examples that use SOM as a feature extractor and then continue to do the further classification or regression predictions with better performance? Many thanks
In this example SOM is used for dimensionality reduction. After applying the model you only have two features, the coordinates on the map.
DeleteUnfortunately I don't have an example that shows what you want to do exactly, but there's an example for classification here https://github.com/JustGlowing/minisom/tree/master/examples
Hi,
ReplyDeleteThank you for your code, it works fantastically! I am performing an analysis on how the random seed changes the resulting neuron weights, and finding that there is indeed some non-negligible variation when using different random seeds, even with PCA initialisation. I've tried to increase the number of iterations, however this does not seem to remedy the problem. What would you suggest to reach a better convergence?
Also, with regards to the sigma value, is it correct that the units for the sigma value would be the same as the units of the output SOM?
Thanks again for some brilliant code!
Hi there,
DeleteRegarding the random seed. It's normal that when you change it, the results change it. SOM is quite sensitive to the initial conditions as most Neural Network but the convergence should not be affected unless you have a very complex problem.
Also, sigma is expressed in grid positions. For example, if you use a sigma of 3 with a bubble neighbourhood function, you'll have a neighbourhood function with a radius of 3.
Hi :) The distance map gives the average distance to the surrounding nodes if I understand correctly. But I'd like the actual distance to each of the nodes (so 4 values for each node on a rectangular grid instead of one averaged value); is there somewhere in the source code that contains this information? I've been looking at the nested for loops in the distance_map function...but am not sure exactly what would need to be returned if it is possible to get the individual distances from that...Thanks in advance for any help you can provide! :)
ReplyDeletehi there, the matrix um at this line of code contains what you are looking for:
Deletehttps://github.com/JustGlowing/minisom/blob/master/minisom.py#L461
The results are averaged only in the last two lines of the method.
Thanks! & thanks for the quick reply :)
DeleteHi, thank you very much for your code! I understand that the activation response shows how often each neuron won. However, I still don't quite understand what the activation map shows yet. Could you explain it? Thanks!
ReplyDeletehi there, the activation map shows the response of each neuron to the input pattern. You can have a look at the source code here: https://github.com/JustGlowing/minisom/blob/master/minisom.py#L237
DeleteThanks for the fast response. So the activation map shows the distance of each neuron to the input data then?
Deleteyes, indeed.
DeleteOkay, thank you very much!
DeleteHi, is there a way to train the SOM for several iterations instead of just one? Thanks!
ReplyDeleteThe method train takes in input the number of iterations to perform.
DeleteCode did not work : Modified som.weights to som._weights and also had to change the target definitions
ReplyDeletefrom pylab import plot,axis,show,pcolor,colorbar,bone
bone()
pcolor(som.distance_map().T) # distance map as background
colorbar()
# loading the labels
target = genfromtxt('iris-dataset.csv', delimiter=',',usecols=(4),dtype=str)
t = zeros(len(target),dtype=int)
t[target=='Iris-setosa'] = 0
t[target=='Iris-versicolor'] = 1
t[target=='Iris-virginica'] = 2
# use different colors and markers for each label
markers = ['o','s','D']
colors = ['r','g','b']
for cnt,xx in enumerate(data):
w = som.winner(xx) # getting the winner
# place a marker on the winning position for the sample xx
plot(w[0]+.5,w[1]+.5,markers[t[cnt]],markerfacecolor='None',
markeredgecolor=colors[t[cnt]],markersize=12,markeredgewidth=2)
axis([0,som._weights.shape[0],0,som._weights.shape[1]])
show() # show the figure
Many many thanks! Great work Sir!