Also this year the Packt Publishing gives me the opportunity to review an interesting book about scientific computing in Python. The last year I had the pleasure to review Numpy 1.5 Beginner's Guide from Ivan Idris and I was surprised about the number of code examples and the straightforward approach to the explanation of the topics. Now I am waiting for Numpy Cookbook, which is from the same author and I can't wait to write the review. Stay tuned ;)
Thursday, December 13, 2012
Thursday, December 6, 2012
3D stem plot
A two-dimensional stem plot displays data as lines extending from a baseline along the x axis. In the following snippet we will see how to make a three-dimensional stem plot using the mplot3d toolkit. In this case we have that data is displayed as lines extending from the x-y plane along the z direction.Let's go with the code:
from numpy import linspace, sin, cos
from pylab import figure, show
from mpl_toolkits.mplot3d import Axes3D
# generating some data
x = linspace(-10,10,100);
y = sin(x);
z = cos(x);
fig = figure()
ax = Axes3D(fig)
# plotting the stems
for i in range(len(x)):
ax.plot([x[i], x[i]], [y[i], y[i]], [0, z[i]],
'--', linewidth=2, color='b', alpha=.5)
# plotting a circle on the top of each stem
ax.plot(x, y, z, 'o', markersize=8,
markerfacecolor='none', color='b',label='ib')
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
show()
And this is the result
Saturday, November 17, 2012
First steps with networkx
One of my favorite topics is the study of structures and, inspired by the presentation of Jacqueline Kazil and Dana Bauer at PyCon US, I started to use networkx in order to analyze some networks. This library provides a lot facilities for the creation, the visualization and the mining of structured data. So, I decided to write this post that shows the first steps to start with it. We will see how to load a network from the gml format and how to prune the network in order to visualize only the nodes with a high degree. In the following examples the coappearance network of characters in the novel Les Miserables, freely available here, will be used. In this network each node represents a character and the connection between two characters represent the coappearance in the same chapter.
Let's start with the snippets. We can load and visualize the network with the following code:
It's easy to see that the graph is not really helpful. Most of the details of the network are still hidden and it's impossible to understand which are the most important nodes. Let's plot an histogram of the number of connections per node:
Looking at this histogram we can see that only few characters have more than ten connections. Then, we decide to visualize only them:
Let's start with the snippets. We can load and visualize the network with the following code:
# read the graph (gml format)
G = nx.read_gml('lesmiserables.gml',relabel=True)
# drawing the full network
figure(1)
nx.draw_spring(G,node_size=0,edge_color='b',alpha=.2,font_size=10)
show()
This should be the result:
It's easy to see that the graph is not really helpful. Most of the details of the network are still hidden and it's impossible to understand which are the most important nodes. Let's plot an histogram of the number of connections per node:
# distribution of the degree figure(2) d = nx.degree(G) hist(d.values(),bins=15) show()The result should be as follows:
Looking at this histogram we can see that only few characters have more than ten connections. Then, we decide to visualize only them:
def trim_nodes(G,d):
""" returns a copy of G without
the nodes with a degree less than d """
Gt = G.copy()
dn = nx.degree(Gt)
for n in Gt.nodes():
if dn[n] <= d:
Gt.remove_node(n)
return Gt
# drawing the network without
# nodes with degree less than 10
Gt = trim_nodes(G,10)
figure(3)
nx.draw(Gt,node_size=0,node_color='w',edge_color='b',alpha=.2)
show()
In the graph below we can see the final result of the analysis. This time the graph makes us able to observe which are the most relevant characters and how they are related to each other according to their coappearance through the chapters.
Saturday, November 3, 2012
Text to Speech with correct intonation
Google has an unoffciale text to speech API. It can be accessed by http requests but it is limited to strings with less than 100 characters. In this post we will see how to split a text longer than 100 characters in order to obtain a correct voice intonation with this service.
The approach is straighforward, we split the text in sentences with less than 100 characters according to the punctuation. Let's see how:
def parseText(text):
""" returns a list of sentences with less than 100 caracters """
toSay = []
punct = [',',':',';','.','?','!'] # punctuation
words = text.split(' ')
sentence = ''
for w in words:
if w[len(w)-1] in punct: # encountered a punctuation mark
if (len(sentence)+len(w)+1 < 100): # is there enough space?
sentence += ' '+w # add the word
toSay.append(sentence.strip()) # save the sentence
else:
toSay.append(sentence.strip()) # save the sentence
toSay.append(w.strip()) # save the word as a sentence
sentence = '' # start another sentence
else:
if (len(sentence)+len(w)+1 < 100):
sentence += ' '+w # add the word
else:
toSay.append(sentence.strip()) # save the sentence
sentence = w # start a new sentence
if len(sentence) > 0:
toSay.append(sentence.strip())
return toSay
Now, we can obtain the speech with a http request for each setence:
text = 'Think of color, pitch, loudness, heaviness, and hotness. Each is the topic of a branch of physics.'
print text
toSay = parseText(text)
google_translate_url = 'http://translate.google.com/translate_tts'
opener = urllib2.build_opener()
opener.addheaders = [('User-agent', 'Mozilla/4.0 (compatible; MSIE 6.0; Windows NT 5.0)')]
for i,sentence in enumerate(toSay):
print i,len(sentence), sentence
response = opener.open(google_translate_url+'?q='+sentence.replace(' ','%20')+'&tl=en')
ofp = open(str(i)+'speech_google.mp3','wb')
ofp.write(response.read())
ofp.close()
os.system('cvlc --play-and-exit -q '+str(i)+'speech_google.mp3')
The API returns the speech using the mp3 format. The code above saves the result of the query and plays it using vlc.
Friday, October 12, 2012
Visualizing correlation matrices
The correlation is one of the most common and most useful statistics. A correlation is a single number that describes the degree of relationship between two variables.
The function corrcoef provided by numpy returns a matrix R of correlation coefficients calculated from an input matrix X whose rows are variables and whose columns are observations. Each element of the matrix R represents the correlation between two variables and it is computed as
where cov(X,Y) is the covariance between X and Y, while σX and σY are the standard deviations. If N is number of variables then R is a N-by-N matrix. Then, when we have a large number of variables we need a way to visualize R. The following snippet uses a pseudocolor plot to visualize R:
As we expected, the correlation coefficients for the variable 2 are higher than the others and we observe a strong correlation between the variables 4 and 8.
where cov(X,Y) is the covariance between X and Y, while σX and σY are the standard deviations. If N is number of variables then R is a N-by-N matrix. Then, when we have a large number of variables we need a way to visualize R. The following snippet uses a pseudocolor plot to visualize R:
from numpy import corrcoef, sum, log, arange from numpy.random import rand from pylab import pcolor, show, colorbar, xticks, yticks # generating some uncorrelated data data = rand(10,100) # each row of represents a variable # creating correlation between the variables # variable 2 is correlated with all the other variables data[2,:] = sum(data,0) # variable 4 is correlated with variable 8 data[4,:] = log(data[8,:])*0.5 # plotting the correlation matrix R = corrcoef(data) pcolor(R) colorbar() yticks(arange(0.5,10.5),range(0,10)) xticks(arange(0.5,10.5),range(0,10)) show()The result should be as follows:
As we expected, the correlation coefficients for the variable 2 are higher than the others and we observe a strong correlation between the variables 4 and 8.
Saturday, September 29, 2012
Weighted random choice
Weighted random choice makes you able to select a random value out of a set of values using a distribution specified though a set of weights. So, given a list we want to pick randomly some elements from it but we need that the chances to pick a specific element is defined using a weight. In the following code we have a function that implements the weighted random choice mechanism and an example of how to use it:
We can observe that the chance to pick the element i is proportional to the weight w[i].
from numpy import cumsum, sort, sum, searchsorted from numpy.random import rand from pylab import hist,show,xticks def weighted_pick(weights,n_picks): """ Weighted random selection returns n_picks random indexes. the chance to pick the index i is give by the weight weights[i]. """ t = cumsum(weights) s = sum(weights) return searchsorted(t,rand(n_picks)*s) # weights, don't have to sum up to one w = [0.1, 0.2, 0.5, 0.5, 1.0, 1.1, 2.0] # picking 10000 times picked_list = weighted_pick(w,10000) # plotting the histogram hist(picked_list,bins=len(w),normed=1,alpha=.8,color='red') show()The code above plots the distribution of the selected indexes:
We can observe that the chance to pick the element i is proportional to the weight w[i].
Friday, September 14, 2012
Boxplot with matplotlib
A boxplot (also known as a box-and-whisker diagram) is a way of summarizing a set of data measured on an interval scale. In this post I will show how to make a boxplot with pylab using a dataset that contains the monthly totals of the number of new cases of measles, mumps, and chicken pox for New York City during the years 1931-1971. The data was extracted from the Hipel-McLeod Time Series Datasets Collection and you can download it from here in the matlab format.
Let's make a box plot of the monthly distribution of chicken pox cases:
On each box, the central mark is the median, the edges of the box are the lower hinge (defined as the 25th percentile) and the upper hinge (the 75th percentile), the whiskers extend to the most extreme data points not considered outliers, these ones are plotted individually.
Using the graph we can compare the range and distribution of the chickenpox cases for each month. We can observe that March and April are the month with the highest number of cases but also the ones with the greatest variability. We can compare the distribution of the three diseases in the same way:
Here, we can observe that the chicken pox distribution has the median higher than the other diseases. The mumps distribution seems to have small variability compared to the other ones and the measles distribution has a low median but a very high number of outliers.
Let's make a box plot of the monthly distribution of chicken pox cases:
from pylab import *
from scipy.io import loadmat
NYCdiseases = loadmat('NYCDiseases.mat') # it's a matlab file
# multiple box plots on one figure
# Chickenpox cases by month
figure(1)
# NYCdiseases['chickenPox'] is a matrix
# with 30 rows (1 per year) and 12 columns (1 per month)
boxplot(NYCdiseases['chickenPox'])
labels = ('Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun',
'Jul', 'Aug', 'Sep', 'Oct', 'Nov', 'Dec')
xticks(range(1,13),labels, rotation=15)
xlabel('Month')
ylabel('Chickenpox cases')
title('Chickenpox cases in NYC 1931-1971')
The result should be as follows:
On each box, the central mark is the median, the edges of the box are the lower hinge (defined as the 25th percentile) and the upper hinge (the 75th percentile), the whiskers extend to the most extreme data points not considered outliers, these ones are plotted individually.
Using the graph we can compare the range and distribution of the chickenpox cases for each month. We can observe that March and April are the month with the highest number of cases but also the ones with the greatest variability. We can compare the distribution of the three diseases in the same way:
# building the data matrix
data = [NYCdiseases['measles'],
NYCdiseases['mumps'], NYCdiseases['chickenPox']]
figure(2)
boxplot(data)
xticks([1,2,3],('measles','mumps','chickenPox'), rotation=15)
ylabel('Monthly cases')
title('Contagious childhood disease in NYC 1931-1971')
show()
And this is the result:
Here, we can observe that the chicken pox distribution has the median higher than the other diseases. The mumps distribution seems to have small variability compared to the other ones and the measles distribution has a low median but a very high number of outliers.
Thursday, August 16, 2012
Kernel Density Estimation with scipy
This post continues the last one where we have seen how to how to fit two types of distribution functions (Normal and Rayleigh). This time we will see how to use Kernel Density Estimation (KDE) to estimate the probability density function. KDE is a non-parametric technique for density estimation in which a known density function (the kernel) is averaged across the observed data points to create a smooth approximation. Given the non-parametrica nature of KDE, the main estimator has not a fixed functional form but only it depends upon all the data points we used for the estimation. Let's see the snippet:
from scipy.stats.kde import gaussian_kde from scipy.stats import norm from numpy import linspace,hstack from pylab import plot,show,hist # creating data with two peaks sampD1 = norm.rvs(loc=-1.0,scale=1,size=300) sampD2 = norm.rvs(loc=2.0,scale=0.5,size=300) samp = hstack([sampD1,sampD2]) # obtaining the pdf (my_pdf is a function!) my_pdf = gaussian_kde(samp) # plotting the result x = linspace(-5,5,100) plot(x,my_pdf(x),'r') # distribution function hist(samp,normed=1,alpha=.3) # histogram show()The result should be as follows:
Friday, July 20, 2012
Distribution fitting with scipy
Distribution fitting is the procedure of selecting a statistical distribution that best fits to a dataset generated by some random process. In this post we will see how to fit a distribution using the techniques implemented in the Scipy library.
This is the first snippet:
In the code above a dataset of 150 samples have been created using a normal distribution with mean 0 and standar deviation 1, then a fitting procedure have been applied on the data. In the figure we can see the original distribution (blue curve) and the fitted distribution (red curve) and we can observe that they are really similar. Let's do the same with a Rayleigh distribution:
As expected, the two distributions are very close.
This is the first snippet:
from scipy.stats import norm
from numpy import linspace
from pylab import plot,show,hist,figure,title
# picking 150 of from a normal distrubution
# with mean 0 and standard deviation 1
samp = norm.rvs(loc=0,scale=1,size=150)
param = norm.fit(samp) # distribution fitting
# now, param[0] and param[1] are the mean and
# the standard deviation of the fitted distribution
x = linspace(-5,5,100)
# fitted distribution
pdf_fitted = norm.pdf(x,loc=param[0],scale=param[1])
# original distribution
pdf = norm.pdf(x)
title('Normal distribution')
plot(x,pdf_fitted,'r-',x,pdf,'b-')
hist(samp,normed=1,alpha=.3)
show()
The result should be as follows
In the code above a dataset of 150 samples have been created using a normal distribution with mean 0 and standar deviation 1, then a fitting procedure have been applied on the data. In the figure we can see the original distribution (blue curve) and the fitted distribution (red curve) and we can observe that they are really similar. Let's do the same with a Rayleigh distribution:
from scipy.stats import norm,rayleigh
samp = rayleigh.rvs(loc=5,scale=2,size=150) # samples generation
param = rayleigh.fit(samp) # distribution fitting
x = linspace(5,13,100)
# fitted distribution
pdf_fitted = rayleigh.pdf(x,loc=param[0],scale=param[1])
# original distribution
pdf = rayleigh.pdf(x,loc=5,scale=2)
title('Rayleigh distribution')
plot(x,pdf_fitted,'r-',x,pdf,'b-')
hist(samp,normed=1,alpha=.3)
show()
The resulting plot:
As expected, the two distributions are very close.
Sunday, July 8, 2012
Color quantization
The aim of color clustering is to produce a small set of representative colors which captures the color properties of an image. Using the small set of color found by the clustering, a quantization process can be applied to the image to find a new version of the image that has been "simplified," both in colors and shapes.
In this post we will see how to use the K-Means algorithm to perform color clustering and how to apply the quantization. Let's see the code:
We have the original image on the top and the quantized version on the bottom. We can see that the image on the bottom has only six colors. Now, we can plot the colors found with the clustering in the RGB space with the following code:
In this post we will see how to use the K-Means algorithm to perform color clustering and how to apply the quantization. Let's see the code:
from pylab import imread,imshow,figure,show,subplot
from numpy import reshape,uint8,flipud
from scipy.cluster.vq import kmeans,vq
img = imread('clearsky.jpg')
# reshaping the pixels matrix
pixel = reshape(img,(img.shape[0]*img.shape[1],3))
# performing the clustering
centroids,_ = kmeans(pixel,6) # six colors will be found
# quantization
qnt,_ = vq(pixel,centroids)
# reshaping the result of the quantization
centers_idx = reshape(qnt,(img.shape[0],img.shape[1]))
clustered = centroids[centers_idx]
figure(1)
subplot(211)
imshow(flipud(img))
subplot(212)
imshow(flipud(clustered))
show()
The result shoud be as follows:
We have the original image on the top and the quantized version on the bottom. We can see that the image on the bottom has only six colors. Now, we can plot the colors found with the clustering in the RGB space with the following code:
# visualizing the centroids into the RGB space from mpl_toolkits.mplot3d import Axes3D fig = figure(2) ax = fig.gca(projection='3d') ax.scatter(centroids[:,0],centroids[:,1],centroids[:,2],c=centroids/255.,s=100) show()And this is the result:
Monday, May 14, 2012
Manifold learning on handwritten digits with Isomap
The Isomap algorithm is an approach to manifold learning. Isomap seeks a lower dimensional embedding of a set of high dimensional data points estimating the intrinsic geometry of a data manifold based on a rough estimate of each data point’s neighbors.
The scikit-learn library provides a great implmentation of the Isomap algorithm and a dataset of handwritten digits. In this post we'll see how to load the dataset and how to compute an embedding of the dataset on a bidimentional space.
Let's load the dataset and show some samples:
Now X is a matrix where each row is a vector that represent a digit. Each vector has 64 elements and it has been obtained using spatial resampling on the above images. We can apply the Isomap algorithm on this data and plot the result with the following lines:
We computed a bidimensional version of each pattern in the dataset and it's easy to see that the separation between the five classes in the new manifold is pretty neat.
The scikit-learn library provides a great implmentation of the Isomap algorithm and a dataset of handwritten digits. In this post we'll see how to load the dataset and how to compute an embedding of the dataset on a bidimentional space.
Let's load the dataset and show some samples:
from pylab import scatter,text,show,cm,figure from pylab import subplot,imshow,NullLocator from sklearn import manifold, datasets # load the digits dataset # 901 samples, about 180 samples per class # the digits represented 0,1,2,3,4 digits = datasets.load_digits(n_class=5) X = digits.data color = digits.target # shows some digits figure(1) for i in range(36): ax = subplot(6,6,i) ax.xaxis.set_major_locator(NullLocator()) # remove ticks ax.yaxis.set_major_locator(NullLocator()) imshow(digits.images[i], cmap=cm.gray_r)The result should be as follows:
Now X is a matrix where each row is a vector that represent a digit. Each vector has 64 elements and it has been obtained using spatial resampling on the above images. We can apply the Isomap algorithm on this data and plot the result with the following lines:
# running Isomap
# 5 neighbours will be considered and reduction on a 2d space
Y = manifold.Isomap(5, 2).fit_transform(X)
# plotting the result
figure(2)
scatter(Y[:,0], Y[:,1], c='k', alpha=0.3, s=10)
for i in range(Y.shape[0]):
text(Y[i, 0], Y[i, 1], str(color[i]),
color=cm.Dark2(color[i] / 5.),
fontdict={'weight': 'bold', 'size': 11})
show()
The new embedding for the data will be as follows:
We computed a bidimensional version of each pattern in the dataset and it's easy to see that the separation between the five classes in the new manifold is pretty neat.
Friday, May 4, 2012
Analyzing your Gmail with Matplotlib
Lately, I read this post about using Mathematica to analyze a Gmail account. I found it very interesting and I worked a with imaplib and matplotlib to create two of the graph they showed:
We can analyze the outgoing mails just using selecting the folder '[Gmail]/Sent Mail':
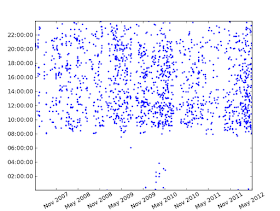
- A diurnal plot, which shows the date and time each email was sent (or received), with years running along the x axis and times of day on the y axis.
- And a daily distribution histogram, which represents the distribution of emails sent by time of day.
from imaplib import IMAP4_SSL
from datetime import date,timedelta,datetime
from time import mktime
from email.utils import parsedate
from pylab import plot_date,show,xticks,date2num
from pylab import figure,hist,num2date
from matplotlib.dates import DateFormatter
def getHeaders(address,password,folder,d):
""" retrieve the headers of the emails
from d days ago until now """
# imap connection
mail = IMAP4_SSL('imap.gmail.com')
mail.login(address,password)
mail.select(folder)
# retrieving the uids
interval = (date.today() - timedelta(d)).strftime("%d-%b-%Y")
result, data = mail.uid('search', None,
'(SENTSINCE {date})'.format(date=interval))
# retrieving the headers
result, data = mail.uid('fetch', data[0].replace(' ',','),
'(BODY[HEADER.FIELDS (DATE)])')
mail.close()
mail.logout()
return data
The second one, make us able to make the diurnal plot:
def diurnalPlot(headers):
""" diurnal plot of the emails,
with years running along the x axis
and times of day on the y axis.
"""
xday = []
ytime = []
for h in headers:
if len(h) > 1:
timestamp = mktime(parsedate(h[1][5:].replace('.',':')))
mailstamp = datetime.fromtimestamp(timestamp)
xday.append(mailstamp)
# Time the email is arrived
# Note that years, month and day are not important here.
y = datetime(2010,10,14,
mailstamp.hour, mailstamp.minute, mailstamp.second)
ytime.append(y)
plot_date(xday,ytime,'.',alpha=.7)
xticks(rotation=30)
return xday,ytime
And this is the function for the daily distribution histogram:
def dailyDistributioPlot(ytime):
""" draw the histogram of the daily distribution """
# converting dates to numbers
numtime = [date2num(t) for t in ytime]
# plotting the histogram
ax = figure().gca()
_, _, patches = hist(numtime, bins=24,alpha=.5)
# adding the labels for the x axis
tks = [num2date(p.get_x()) for p in patches]
xticks(tks,rotation=75)
# formatting the dates on the x axis
ax.xaxis.set_major_formatter(DateFormatter('%H:%M'))
Now we got everything we need to make the graphs. Let's try to analyze the outgoing mails of last 5 years:
print 'Fetching emails...'
headers = getHeaders('iamsupersexy@gmail.com',
'ofcourseiamsupersexy','inbox',365*5)
print 'Plotting some statistics...'
xday,ytime = diurnalPlot(headers)
dailyDistributioPlot(ytime)
print len(xday),'Emails analysed.'
show()
The result would appear as follows
We can analyze the outgoing mails just using selecting the folder '[Gmail]/Sent Mail':
print 'Fetching emails...'
headers = getHeaders('iamsupersexy@gmail.com',
'ofcourseiamsupersexy','[Gmail]/Sent Mail',365*5)
print 'Plotting some statistics...'
xday,ytime = diurnalPlot(headers)
dailyDistributioPlot(ytime)
print len(xday),'Emails analysed.'
show()
And this is the result:
Thursday, April 26, 2012
K-Nearest Neighbour Classifier
The Nearest Neighbour Classifier is one of the most straightforward classifier in the arsenal of machine learning techniques. It performs the classification by identifying the nearest neighbours to a query pattern and using those neighbors to determine the label of the query. The idea behind the algorithm is simple: Assign the query pattern to the class which occurs the most in the k nearest neighbors. In this post we'll use the function knn_search(...) that we have seen in the last post to implement a K-Nearest Neighbour Classifier. The implementation of the classifier is as follows:
The query vector is represented with a green point and we can see that the 3 out of 5 nearest neighbors are red points (label 1) while the remaining 2 are blue (label 2).
The result of the classification will be printed on the console:
from numpy import random,argsort,argmax,bincount,int_,array,vstack,round
from pylab import scatter,show
def knn_classifier(x, D, labels, K):
""" Classify the vector x
D - data matrix (each row is a pattern).
labels - class of each pattern.
K - number of neighbour to use.
Returns the class label and the neighbors indexes.
"""
neig_idx = knn_search(x,D,K)
counts = bincount(labels[neig_idx]) # voting
return argmax(counts),neig_idx
Let's test the classifier on some random data:
# generating a random dataset with random labels data = random.rand(2,150) # random points labels = int_(round(random.rand(150)*1)) # random labels 0 or 1 x = random.rand(2,1) # random test point # label assignment using k=5 result,neig_idx = knn_classifier(x,data,labels,5) print 'Label assignment:', result # plotting the data and the input pattern # class 1, red points, class 0 blue points scatter(data[0,:],data[1,:], c=labels,alpha=0.8) scatter(x[0],x[1],marker='o',c='g',s=40) # highlighting the neighbours plot(data[0,neig_idx],data[1,neig_idx],'o', markerfacecolor='None',markersize=15,markeredgewidth=1) show()The script will show the following graph:
The query vector is represented with a green point and we can see that the 3 out of 5 nearest neighbors are red points (label 1) while the remaining 2 are blue (label 2).
The result of the classification will be printed on the console:
Label assignment: 1As we expected, the green point have been assigned to the class with red markers.
Saturday, April 14, 2012
k-nearest neighbor search
A k-nearest neighbor search identifies the top k nearest neighbors to a query. The problem is: given a dataset D of vectors in a d-dimensional space and a query point x in the same space, find the closest point in D to x. The following function performs a k-nearest neighbor search using the euclidean distance:
Now, we will test this function on a random bidimensional dataset:
The red point is the query vector and the blue ones represent the data. The blue points surrounded by a black circle are the nearest neighbors.
from numpy import random,argsort,sqrt from pylab import plot,show def knn_search(x, D, K): """ find K nearest neighbours of data among D """ ndata = D.shape[1] K = K if K < ndata else ndata # euclidean distances from the other points sqd = sqrt(((D - x[:,:ndata])**2).sum(axis=0)) idx = argsort(sqd) # sorting # return the indexes of K nearest neighbours return idx[:K]The function computes the euclidean distance between every point of D and x then returns the indexes of the points for which the distance is smaller.
Now, we will test this function on a random bidimensional dataset:
# knn_search test data = random.rand(2,200) # random dataset x = random.rand(2,1) # query point # performing the search neig_idx = knn_search(x,data,10) # plotting the data and the input point plot(data[0,:],data[1,:],'ob',x[0,0],x[1,0],'or') # highlighting the neighbours plot(data[0,neig_idx],data[1,neig_idx],'o', markerfacecolor='None',markersize=15,markeredgewidth=1) show()The result is as follows:
The red point is the query vector and the blue ones represent the data. The blue points surrounded by a black circle are the nearest neighbors.
Thursday, April 5, 2012
K- means clustering with scipy
K-means clustering is a method for finding clusters and cluster centers in a set of unlabeled data. Intuitively, we might think of a cluster as comprising a group of data points whose inter-point distances are small compared with the distances to points outside of the cluster. Given an initial set of K centers, the K-means algorithm alternates the two steps:
The Scipy library provides a good implementation of the K-Means algorithm. Let's see how to use it:
In this case we splitted the data in 2 clusters, the blue points have been assigned to the first and the red ones to the second. The squares are the centers of the clusters.
Let's see try to split the data in 3 clusters:
- for each center we identify the subset of training points (its cluster) that is closer to it than any other center;
- the means of each feature for the data points in each cluster are computed, and this mean vector becomes the new center for that cluster.
The Scipy library provides a good implementation of the K-Means algorithm. Let's see how to use it:
from pylab import plot,show
from numpy import vstack,array
from numpy.random import rand
from scipy.cluster.vq import kmeans,vq
# data generation
data = vstack((rand(150,2) + array([.5,.5]),rand(150,2)))
# computing K-Means with K = 2 (2 clusters)
centroids,_ = kmeans(data,2)
# assign each sample to a cluster
idx,_ = vq(data,centroids)
# some plotting using numpy's logical indexing
plot(data[idx==0,0],data[idx==0,1],'ob',
data[idx==1,0],data[idx==1,1],'or')
plot(centroids[:,0],centroids[:,1],'sg',markersize=8)
show()
The result should be as follows:
In this case we splitted the data in 2 clusters, the blue points have been assigned to the first and the red ones to the second. The squares are the centers of the clusters.
Let's see try to split the data in 3 clusters:
# now with K = 3 (3 clusters)
centroids,_ = kmeans(data,3)
idx,_ = vq(data,centroids)
plot(data[idx==0,0],data[idx==0,1],'ob',
data[idx==1,0],data[idx==1,1],'or',
data[idx==2,0],data[idx==2,1],'og') # third cluster points
plot(centroids[:,0],centroids[:,1],'sm',markersize=8)
show()
This time the the result is as follows:
Saturday, March 24, 2012
Linear regression with Numpy
Few post ago, we have seen how to use the function numpy.linalg.lstsq(...) to solve an over-determined system. This time, we'll use it to estimate the parameters of a regression line.
A linear regression line is of the form w1x+w2=y and it is the line that minimizes the sum of the squares of the distance from each data point to the line. So, given n pairs of data (xi, yi), the parameters that we are looking for are w1 and w2 which minimize the error
and we can compute the parameter vector w = (w1 , w2)T as the least-squares solution of the following over-determined system
Let's use numpy to compute the regression line:
You can find more about data fitting using numpy in the following posts: Update, the same result could be achieve using the function scipy.stats.linregress (thanks ianalis!):
A linear regression line is of the form w1x+w2=y and it is the line that minimizes the sum of the squares of the distance from each data point to the line. So, given n pairs of data (xi, yi), the parameters that we are looking for are w1 and w2 which minimize the error
and we can compute the parameter vector w = (w1 , w2)T as the least-squares solution of the following over-determined system
Let's use numpy to compute the regression line:
from numpy import arange,array,ones,linalg from pylab import plot,show xi = arange(0,9) A = array([ xi, ones(9)]) # linearly generated sequence y = [19, 20, 20.5, 21.5, 22, 23, 23, 25.5, 24] w = linalg.lstsq(A.T,y)[0] # obtaining the parameters # plotting the line line = w[0]*xi+w[1] # regression line plot(xi,line,'r-',xi,y,'o') show()We can see the result in the plot below.
You can find more about data fitting using numpy in the following posts: Update, the same result could be achieve using the function scipy.stats.linregress (thanks ianalis!):
from numpy import arange,array,ones#,random,linalg from pylab import plot,show from scipy import stats xi = arange(0,9) A = array([ xi, ones(9)]) # linearly generated sequence y = [19, 20, 20.5, 21.5, 22, 23, 23, 25.5, 24] slope, intercept, r_value, p_value, std_err = stats.linregress(xi,y) print 'r value', r_value print 'p_value', p_value print 'standard deviation', std_err line = slope*xi+intercept plot(xi,line,'r-',xi,y,'o') show()
Friday, March 16, 2012
SVD decomposition with numpy
The SVD decomposition is a factorization of a matrix, with many useful applications in signal processing and statistics. In this post we will see
Since U and V are orthogonal (this means that U^T*U=I and V^T*V=I) we can write the inverse of A as (see Solving overdetermined systems with the QR decomposition for the tricks)
So, let's start computing the factorization and the inverse
or solving the system
Multiplying by U^T we obtain
Then, letting c be U^Tb and w be V^Tx, we see
Since sigma is diagonal, we can easily obtain w solving the system above. And finally, we can obtain x solving
Let's compare the results of those methods:
- how to compute the SVD decomposition of a matrix A using numpy,
- how to compute the inverse of A using the matrices computed by the decomposition,
- and how to solve a linear equation system Ax=b using using the SVD.
Since U and V are orthogonal (this means that U^T*U=I and V^T*V=I) we can write the inverse of A as (see Solving overdetermined systems with the QR decomposition for the tricks)
So, let's start computing the factorization and the inverse
from numpy import * A = floor(random.rand(4,4)*20-10) # generating a random b = floor(random.rand(4,1)*20-10) # system Ax=b U,s,V = linalg.svd(A) # SVD decomposition of A # computing the inverse using pinv pinv = linalg.pinv(A) # computing the inverse using the SVD decomposition pinv_svd = dot(dot(V.T,linalg.inv(diag(s))),U.T) print "Inverse computed by lingal.pinv()\n",pinv print "Inverse computed using SVD\n",pinv_svdAs we can see, the output shows that pinv and pinv_svd are the equal
Inverse computed by lingal.pinv() [[ 0.06578301 -0.04663721 0.0436917 0.089838 ] [ 0.15243004 0.044919 -0.03681885 0.00294551] [ 0.18213058 -0.01718213 0.06872852 -0.07216495] [ 0.03976436 0.09867452 0.03387334 -0.04270987]] Inverse computed using SVD [[ 0.06578301 -0.04663721 0.0436917 0.089838 ] [ 0.15243004 0.044919 -0.03681885 0.00294551] [ 0.18213058 -0.01718213 0.06872852 -0.07216495] [ 0.03976436 0.09867452 0.03387334 -0.04270987]]Now, we can solve Ax=b using the inverse:
or solving the system
Multiplying by U^T we obtain
Then, letting c be U^Tb and w be V^Tx, we see
Since sigma is diagonal, we can easily obtain w solving the system above. And finally, we can obtain x solving
Let's compare the results of those methods:
x = linalg.solve(A,b) # solve Ax=b using linalg.solve xPinv = dot(pinv_svd,b) # solving Ax=b computing x = A^-1*b # solving Ax=b using the equation above c = dot(U.T,b) # c = U^t*b w = linalg.solve(diag(s),c) # w = V^t*c xSVD = dot(V.T,w) # x = V*w print "Ax=b solutions compared" print x.T print xSVD.T print xPinv.TAs aspected, we have the same solutions:
Ax=b solutions compared [[ 0.13549337 -0.37260677 1.62886598 -0.09720177]] [[ 0.13549337 -0.37260677 1.62886598 -0.09720177]] [[ 0.13549337 -0.37260677 1.62886598 -0.09720177]]
Saturday, March 10, 2012
Solving overdetermined systems with the QR decomposition
A system of linear equations is considered overdetermined if there are more equations than unknowns. In practice, we have a system Ax=b where A is a m by n matrix and b is a m dimensional vector b but m is greater than n. In this case, the vector b cannot be expressed as a linear combination of the columns of A. Hence, we can't find x so that satisfies the problem Ax=b (except in specific cases) but it is possible to determine x so that Ax is as close to b as possible. So we wish to find x which minimizes the following error
Considering the QR decomposition of A we have that Ax=b becomes
multiplying by Q^T we obtain
and since Q^T is orthogonal (this means that Q^T*Q=I) we have
Now, this is a well defined system, R is an upper triangular matrix and Q^T*b is a vector. More precisely b is the orthogonal projection of b onto the range of A. And,
The function linalg.lstsq() provided by numpy returns the least-squares solution to a linear system equation and is able to solve overdetermined systems. Let's compare the solutions of linalg.lstsq() with the ones computed using the QR decomposition:
Considering the QR decomposition of A we have that Ax=b becomes
multiplying by Q^T we obtain
and since Q^T is orthogonal (this means that Q^T*Q=I) we have
Now, this is a well defined system, R is an upper triangular matrix and Q^T*b is a vector. More precisely b is the orthogonal projection of b onto the range of A. And,
The function linalg.lstsq() provided by numpy returns the least-squares solution to a linear system equation and is able to solve overdetermined systems. Let's compare the solutions of linalg.lstsq() with the ones computed using the QR decomposition:
from numpy import * # generating a random overdetermined system A = random.rand(5,3) b = random.rand(5,1) x_lstsq = linalg.lstsq(A,b)[0] # computing the numpy solution Q,R = linalg.qr(A) # qr decomposition of A Qb = dot(Q.T,b) # computing Q^T*b (project b onto the range of A) x_qr = linalg.solve(R,Qb) # solving R*x = Q^T*b # comparing the solutions print 'qr solution' print x_qr print 'lstqs solution' print x_lstsqThis is the output of the script above:
qr solution [[ 0.08704059] [-0.10106932] [ 0.56961487]] lstqs solution [[ 0.08704059] [-0.10106932] [ 0.56961487]]As we can see, the solutions are the same.
Tuesday, February 28, 2012
Finite differences with Toeplitz matrix
A Toeplitz matrix is a band matrix in which each descending diagonal from left to right is constant. In this post we will see how to approximate the derivative of a function f(x) as matrix-vector products between a Toeplitz matrix and a vector of equally spaced values of f.
Let's see how to generate the matrices we need using the function toeplitz(...) provided by numpy:
Forward difference, Backward difference, And central difference,
where h is the step size between the samples. Those equations are called Finite Differences and they give us an approximate derivative of f. So, let's approximate some derivatives!
As the theory suggests, the approximation is better when h is smaller and the central differences are more accurate (note that, they have a higher order of accuracy respect to the backward and forward ones).
from numpy import * from scipy.linalg import toeplitz import pylab def forward(size): """ returns a toeplitz matrix for forward differences """ r = zeros(size) c = zeros(size) r[0] = -1 r[size-1] = 1 c[1] = 1 return toeplitz(r,c) def backward(size): """ returns a toeplitz matrix for backward differences """ r = zeros(size) c = zeros(size) r[0] = 1 r[size-1] = -1 c[1] = -1 return toeplitz(r,c).T def central(size): """ returns a toeplitz matrix for central differences """ r = zeros(size) c = zeros(size) r[1] = .5 r[size-1] = -.5 c[1] = -.5 c[size-1] = .5 return toeplitz(r,c).T # testing the functions printing some 4-by-4 matrices print 'Forward matrix' print forward(4) print 'Backward matrix' print backward(4) print 'Central matrix' print central(4)The result of the test above is as follows:
Forward matrix [[-1. 1. 0. 0.] [ 0. -1. 1. 0.] [ 0. 0. -1. 1.] [ 1. 0. 0. -1.]] Backward matrix [[ 1. 0. 0. -1.] [-1. 1. 0. 0.] [ 0. -1. 1. 0.] [ 0. 0. -1. 1.]] Central matrix [[ 0. 0.5 0. -0.5] [-0.5 0. 0.5 0. ] [ 0. -0.5 0. 0.5] [ 0.5 0. -0.5 0. ]]We can observe that the matrix-vector product between those matrices and the vector of equally spaced values of f(x) implements, respectively, the following equations:
Forward difference, Backward difference, And central difference,
where h is the step size between the samples. Those equations are called Finite Differences and they give us an approximate derivative of f. So, let's approximate some derivatives!
x = linspace(0,10,15) y = cos(x) # recall, the derivative of cos(x) is sin(x) # we need the step h to compute f'(x) # because the product gives h*f'(x) h = x[1]-x[2] # generating the matrices Tf = forward(15)/h Tb = backward(15)/h Tc = central(15)/h pylab.subplot(211) # approximation and plotting pylab.plot(x,dot(Tf,y),'g',x,dot(Tb,y),'r',x,dot(Tc,y),'m') pylab.plot(x,sin(x),'b--',linewidth=3) pylab.axis([0,10,-1,1]) # the same experiment with more samples (h is smaller) x = linspace(0,10,50) y = cos(x) h = x[1]-x[2] Tf = forward(50)/h Tb = backward(50)/h Tc = central(50)/h pylab.subplot(212) pylab.plot(x,dot(Tf,y),'g',x,dot(Tb,y),'r',x,dot(Tc,y),'m') pylab.plot(x,sin(x),'b--',linewidth=3) pylab.axis([0,10,-1,1]) pylab.legend(['Forward', 'Backward', 'Central', 'True f prime'],loc=4) pylab.show()The resulting plot would appear as follows:
As the theory suggests, the approximation is better when h is smaller and the central differences are more accurate (note that, they have a higher order of accuracy respect to the backward and forward ones).
Sunday, February 5, 2012
Convolution with numpy
A convolution is a way to combine two sequences, x and w, to get a third sequence, y, that is a filtered version of x. The convolution of the sample xt is computed as follows:
It is the mean of the weighted summation over a window of length k and wt are the weights. Usually, the sequence w is generated using a window function. Numpy has a number of window functions already implemented: bartlett, blackman, hamming, hanning and kaiser. So, let's plot some Kaiser windows varying the parameter beta:
And now, we can use the function convolve(...) to compute the convolution between a vector x and one of the Kaiser window we have seen above:
As we can see, the original sequence have been smoothed by the windows.
It is the mean of the weighted summation over a window of length k and wt are the weights. Usually, the sequence w is generated using a window function. Numpy has a number of window functions already implemented: bartlett, blackman, hamming, hanning and kaiser. So, let's plot some Kaiser windows varying the parameter beta:
import numpy
import pylab
beta = [2,4,16,32]
pylab.figure()
for b in beta:
w = numpy.kaiser(101,b)
pylab.plot(range(len(w)),w,label="beta = "+str(b))
pylab.xlabel('n')
pylab.ylabel('W_K')
pylab.legend()
pylab.show()
The graph would appear as follows:
And now, we can use the function convolve(...) to compute the convolution between a vector x and one of the Kaiser window we have seen above:
def smooth(x,beta): """ kaiser window smoothing """ window_len=11 # extending the data at beginning and at the end # to apply the window at the borders s = numpy.r_[x[window_len-1:0:-1],x,x[-1:-window_len:-1]] w = numpy.kaiser(window_len,beta) y = numpy.convolve(w/w.sum(),s,mode='valid') return y[5:len(y)-5]Let's test it on a random sequence:
# random data generation y = numpy.random.random(100)*100 for i in range(100): y[i]=y[i]+i**((150-i)/80.0) # modifies the trend # smoothing the data pylab.figure(1) pylab.plot(y,'-k',label="original signal",alpha=.3) for b in beta: yy = smooth(y,b) pylab.plot(yy,label="filtered (beta = "+str(b)+")") pylab.legend() pylab.show()The program would have an output similar to the following:
As we can see, the original sequence have been smoothed by the windows.
Saturday, January 21, 2012
Monte Carlo estimate for pi with numpy
In this post we will use a Monte Carlo method to approximate pi. The idea behind the method that we are going to see is the following:
Draw the unit square and the unit circle. Consider only the part of the circle inside the square and pick uniformly a large number of points at random over the square. Now, the unit circle has pi/4 the area of the square. So, it should be apparent that of the total number of points that hit within the square, the number of points that hit the circle quadrant is proportional to the area of that part. This gives a way to approximate pi/4 as the ratio between the number of points inside circle and the total number of points and multiplying it by 4 we have pi.
Let's see the python script that implements the method discussed above using the numpy's indexing facilities:
Note that the lines of code used to estimate pi are just 3!
Draw the unit square and the unit circle. Consider only the part of the circle inside the square and pick uniformly a large number of points at random over the square. Now, the unit circle has pi/4 the area of the square. So, it should be apparent that of the total number of points that hit within the square, the number of points that hit the circle quadrant is proportional to the area of that part. This gives a way to approximate pi/4 as the ratio between the number of points inside circle and the total number of points and multiplying it by 4 we have pi.
Let's see the python script that implements the method discussed above using the numpy's indexing facilities:
from pylab import plot,show,axis
from numpy import random,sqrt,pi
# scattering n points over the unit square
n = 1000000
p = random.rand(n,2)
# counting the points inside the unit circle
idx = sqrt(p[:,0]**2+p[:,1]**2) < 1
plot(p[idx,0],p[idx,1],'b.') # point inside
plot(p[idx==False,0],p[idx==False,1],'r.') # point outside
axis([-0.1,1.1,-0.1,1.1])
show()
# estimation of pi
print '%0.16f' % (sum(idx).astype('double')/n*4),'result'
print '%0.16f' % pi,'real pi'
The program will print the pi approximation on the standard out:
3.1457199999999998 result 3.1415926535897931 real piand will show a graph with the generated points:
Note that the lines of code used to estimate pi are just 3!
Saturday, January 14, 2012
How to plot a function of two variables with matplotlib
In this post we will see how to visualize a function of two variables in two ways. First, we will create an intensity image of the function and, second, we will use the 3D plotting capabilities of matplotlib to create a shaded surface plot. So, let's go with the code:
And now we are going to use the values stored in X,Y and Z to make a 3D plot using the mplot3d toolkit. Here's the snippet:
from numpy import exp,arange
from pylab import meshgrid,cm,imshow,contour,clabel,colorbar,axis,title,show
# the function that I'm going to plot
def z_func(x,y):
return (1-(x**2+y**3))*exp(-(x**2+y**2)/2)
x = arange(-3.0,3.0,0.1)
y = arange(-3.0,3.0,0.1)
X,Y = meshgrid(x, y) # grid of point
Z = z_func(X, Y) # evaluation of the function on the grid
im = imshow(Z,cmap=cm.RdBu) # drawing the function
# adding the Contour lines with labels
cset = contour(Z,arange(-1,1.5,0.2),linewidths=2,cmap=cm.Set2)
clabel(cset,inline=True,fmt='%1.1f',fontsize=10)
colorbar(im) # adding the colobar on the right
# latex fashion title
title('$z=(1-x^2+y^3) e^{-(x^2+y^2)/2}$')
show()
The script would have the following output:And now we are going to use the values stored in X,Y and Z to make a 3D plot using the mplot3d toolkit. Here's the snippet:
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
from matplotlib.ticker import LinearLocator, FormatStrFormatter
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.gca(projection='3d')
surf = ax.plot_surface(X, Y, Z, rstride=1, cstride=1,
cmap=cm.RdBu,linewidth=0, antialiased=False)
ax.zaxis.set_major_locator(LinearLocator(10))
ax.zaxis.set_major_formatter(FormatStrFormatter('%.02f'))
fig.colorbar(surf, shrink=0.5, aspect=5)
plt.show()
And this is the result: