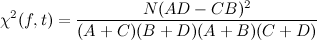To compute these values we need to scrape the career statistics (number of goals and appearances per season) on the Wikipedia pages of the players:
from bs4 import BeautifulSoup
from urllib2 import urlopen
def get_total_goals(url):
"""
Given the url of a wikipedia page about a football striker
returns three numy arrays:
- years, each element corresponds to a season
- apprearances, contains the number of appearances each season
- goals, contains the number of goal scored each season
Unfortunately this function is able to parse
only the pages of few strikers.
"""
soup = BeautifulSoup(urlopen(url).read())
table = soup.find("table", { "class" : "wikitable" })
years = []
apps = []
goals = []
for row in table.findAll("tr"):
cells = row.findAll("td")
if len(cells) > 1:
years.append(int(cells[0].text[:4]))
apps.append(int(cells[len(cells)-2].text))
goals.append(int(cells[len(cells)-1].text))
return np.array(years),
np.array(apps, dtype='float'),
np.array(goals)
ronaldo = get_total_goals('http://en.wikipedia.org/wiki/Ronaldo')
vieri = get_total_goals('http://en.wikipedia.org/wiki/Christian_Vieri')
delpiero = get_total_goals('http://en.wikipedia.org/wiki/Alessandro_Del_Piero')
trezeguet = get_total_goals('http://en.wikipedia.org/wiki/David_Trezeguet')
Now we are ready to compute our statistics. For each statistics we will produce an interactive chart using plotly.
Scoring trajectory
import plotly.plotly as py
from plotly.graph_objs import *
py.sign_in("sexyusername", "mypassword")
data = Data([
Scatter(x=delpiero[0],y=cumsum(delpiero[2]),
name='Del Piero', mode='lines'),
Scatter(x=trezeguet[0],y=cumsum(trezeguet[2]),
name='Trezeguet', mode='lines'),
Scatter(x=ronaldo[0],y=cumsum(ronaldo[2]),
name='Ronaldo', mode='lines'),
Scatter(x=vieri[0],y=cumsum(vieri[2]),
name='Vieri', mode='lines'),
])
layout = Layout(
title='Scoring Trajectory',
xaxis=XAxis(title='Year'),
yaxis=YAxis(title='Cumuative goal'),
legend=Legend(x=0.0,y=1.0))
fig = Figure(data=data, layout=layout)
py.iplot(fig, filename='cumulative-goals')
The scoring trajectory is given by the yearly cumulative totals of goals scored. From the scoring trajectories we can see that Ronaldo was a goal machine since his first professional season and his worse period was from 1999 to 2001. Del Piero and Trezeguet have the longest careers (and they're still playing!). Vieri had the shortest career but it's impressive to see that the number of goals he scored increased almost constantly from 1996 to 2004.
Scoring rate
data = Data([
Bar(
x=['Ronaldo', 'Vieri', 'Trezeguet', 'Del Piero'],
y=[np.sum(ronaldo[2])/np.sum(ronaldo[1]),
np.sum(vieri[2])/np.sum(vieri[1]),
np.sum(trezeguet[2])/np.sum(trezeguet[1]),
np.sum(delpiero[2])/np.sum(delpiero[1])]
)
])
py.iplot(data, filename='goal-average')
The scoring rate is the number of goals scored divided by the number of appearances. Ronaldo has a terrific 0.67 scoring rate, meaning that, on average he scored more than three goals each five games. Vieri and Trezeguet have a very similar scoring rate, almost one goal each two games. While Del Piero has 0.40, two goals each five games.
Appearances
data = Data([
Bar(
x=['Del Piero', 'Trezeguet', 'Ronaldo', 'Vieri'],
y=[np.sum(delpiero[1]),
np.sum(trezeguet[1]),
np.sum(ronaldo[1]),
np.sum(vieri[1])]
)
])
py.iplot(data, filename='appearances')
The number of Del Piero's appearances on a football field is impressive. At the moment I'm writing, he played 773 games. No one of the other players was able to play the 70% of the games played by the Italian numero 10.