Let's define a goal for our Neural Network. We have a dataset H and we want to build a model able to reconstruct the same data. More formally, we want to build a function f that takes in input H and returns the same values or an approximation close as possible to H. We can say that we want f to minimize the following error
To begin our experiment we build a data matrix H that contains the coordinate of a stylized star:
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
x= [np.cos(np.pi/2), 2/5*np.cos(np.pi/2+np.pi/5), np.cos(np.pi/2+2*np.pi/5),
2/5*np.cos(np.pi/2+3*np.pi/5), np.cos(np.pi/2+4*np.pi/5),
2/5*np.cos(3*np.pi/2), np.cos(np.pi/2+6*np.pi/5),
2/5*np.cos(np.pi/2+7*np.pi/5), np.cos(np.pi/2+8*np.pi/5),
2/5*np.cos(np.pi/2+9*np.pi/5), np.cos(np.pi/2)]
y=[np.sin(np.pi/2), 2/5*np.sin(np.pi/2+np.pi/5), np.sin(np.pi/2+2*np.pi/5),
2/5*np.sin(np.pi/2+3*np.pi/5), np.sin(np.pi/2+4*np.pi/5),
2/5*np.sin(3*np.pi/2), np.sin(np.pi/2+6*np.pi/5),
2/5*np.sin(np.pi/2+7*np.pi/5), np.sin(np.pi/2+8*np.pi/5),
2/5*np.sin(np.pi/2+9*np.pi/5), np.sin(np.pi/2)]
xp = np.linspace(0, 1, len(x))
np.interp(2.5, xp, x)
x = np.log(np.interp(np.linspace(0, 1, len(x)*10), xp, x)+1)
yp = np.linspace(0, 1, len(y))
np.interp(2.5, yp, y)
y = np.interp(np.linspace(0, 1, len(y)*10), yp, y)
#y[::2] += .1
H = np.array([x, y]).T
plt.plot(H[:,0], H[:,1], '-o')
Now the matrix H contains the x coordinates of our star in the first column and the y coordinates in the second. With the help of sklearn, we can now train a Neural Network and plot the result:
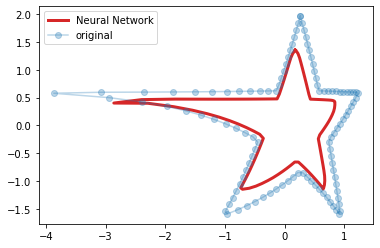
from sklearn.neural_network import MLPRegressor from sklearn.preprocessing import minmax_scale H = scale(H) plt.figure() f = MLPRegressor(hidden_layer_sizes=(200, 200, 200), alpha=0) f.fit(H, H) result = f.predict(H) plt.plot(result[:,0], result[:,1], 'C3', linewidth=3, label='Neural Network') plt.plot(H[:,0], H[:,1], 'o', alpha=.3, label='original') plt.legend() #plt.xlim([-0.1, 1.1]) #plt.ylim([-0.1, 1.1]) plt.show()
In the snippet above we created a Neural Network with 3 layers of 200 neurons. Then, we trained the model using H as both input and output data. In the chart we compare the original data with the estimation. It's easy to see that there are small differences between the two stars but they are very close.
Here's important to notice that we initialized MLPRegressor using alpha=0. This parameter controls the regularization of the model and the higher its value, the more regularization we apply. To understand how alpha affects the learning of the model we need to add a term to the computation of the error that was just introduced:
where W is a matrix of all the weights in the network. The error not only takes in account the difference between the ouput of the function and the data, but also the size of weights of the connections of the network. Hence, the higher alpha, the less the model is free to learn. If we set alpha to 20 in our experiment we have the following result:
We still achie an approximation of the star but the output of our model is smaller than before and the edges of the star are smoother.
Increasing alpha gradually is a good way to understand the effects of regularization:
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
fig, ax = plt.subplots()
ln, = plt.plot([], [], 'C3', linewidth=3, label='Neural Network')
plt.plot(H[:,0], H[:,1], 'o', alpha=.3, label='original')
plt.legend()
def update(frame):
f = MLPRegressor(hidden_layer_sizes=(200, 200, 200), alpha=frame)
f.fit(H, H)
result = f.predict(H)
ln.set_data(result[:,0], result[:,1])
plt.title('alpha = %.2f' % frame)
return ln,
ani = FuncAnimation(fig, update, frames=np.linspace(0, 40, 100), blit=True)
HTML(ani.to_html5_video())
Here we vary alpha from 0 to 40 and plot the result for each value. We notice here that not only the star gets smaller and smoother as alpha increases but also that the network tends to preserve long lines as much as possible getting away from the edges which account less on error function. Finally we see that the result degenerates into a point when alpha is too high.